Yesterday I posted an example of the Pima dataset which provide data on the features of an individual and their likelihood to be diabetic. I didn’t get great results (only 67%), so I wanted to take another look and see if there was anything that I could change to make it better. The dataset is pretty small, only 768 records. In my readings, it showed that when you have a small population of data you can use K-Fold Cross Validation to improve the performance of the model.
K-Fold splits the data into k folds or groups. For instance if you set k to be 3, the data will be split into 1 validation set and 2 training sets. For each k the 2 training sets will be used for… fitting the model and the remaining will be used for validation. SciKitLearn has a KFold objects that you can use to parcel the data into the sets. An interesting point that I didn’t catch at first is that the split function returns sets of indices, not a new list of data.
for train_index, test_index in kf.split(x_data,y_data):
So now that we have our data split into groups, we need to loop over those groups to train the model. Remembering that the split data is just an array of indices, we need to populate our training and test data.
X_train, X_test = x_data[train_index], x_data[test_index]
y_train, y_split_test = y_data[train_index], y_data[test_index]
Just like the previous Pima example, we build, compile, fit and evaluate our model.
model = models.Sequential()
model.add(layers.Dense(16, activation='relu', input_shape=(8,)))
model.add(layers.Dense(16, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
model.compile(optimizer='rmsprop', loss='binary_crossentropy',
metrics=['accuracy'])
history = model.fit(X_train, y_train, epochs=epochs, batch_size=15 )
results = model.evaluate(X_test, y_split_test)
Now to capture the metrics of each fold, we need to store them in an array and I set aside the model with the best performance.
current = results[1]
m = 0
if(len(accuracy_per_fold) > 0):
m = max(accuracy_per_fold)
if current > m :
best_model = model
chosen_model = pass_index
loss_per_fold.append(results[0])
accuracy_per_fold.append(results[1])
Putting it all together after all folds have been processed, we can print the results.
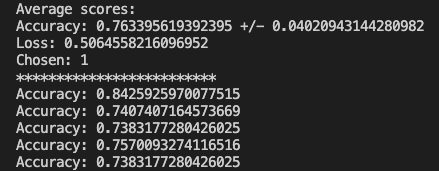
Now we can run our test data through the model and check out the results.
y_new = best_model.predict_classes(x_test)
total = len(y_new)
correct = 0
for i in range(len(accuracy_per_fold)):
print(f'Accuracy: {accuracy_per_fold[i]}')
for i in range(len(x_test)):
if y_test[i] == y_new[i]:
correct +=1
print(correct / total)
Everything worked and based on the randomized test data I was able to achieve a 75% accuracy where the previous method yielded 67%. The full code can be found on GitHub. Just like the other posts, these are just my learnings from the book Deep Learning with Python from Francois Chollet. If any expert reads through this and there is something that I missed or was found to be incorrect, please drop a comment. I am learning so any correction would be appreciated.